CRISPR-CAS9 as a tool for IT-15 gene editing in Huntington's Disease
Main Article Content
Abstract
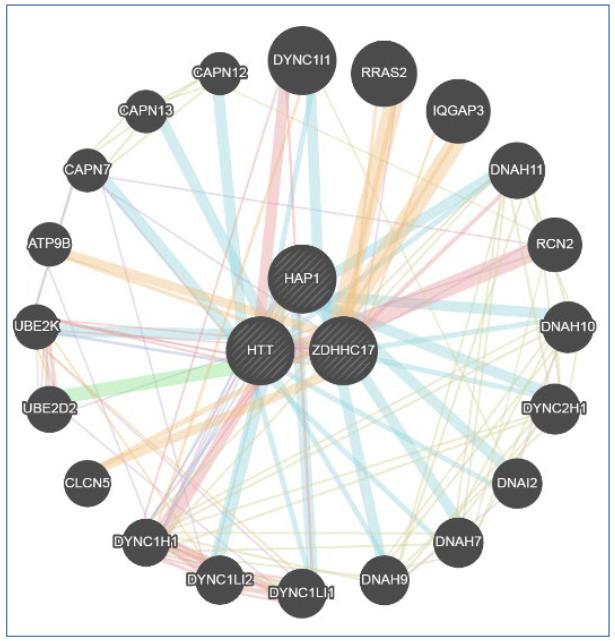
Huntington's disease (HD) is a neurodegenerative, autosomal dominant and hereditary disease that occurs due to a genetic mutation that generates a repetitive sequence of CAG trinucleotides present in the IT-15 gene, huntingtin gene, located on chromosome 4. The objective was to review Huntington's disease (HD) neuropathology and the CRISPR-Cas9 method to silence the IT-15 gene and thus verify the consequence in the HIP14 and HAP1 genes, which have interaction with the mutated huntingtin and the result of this in the patient's organism. Articles were searched in indexed databases (Scielo, PubMed and LILACs) with the following descriptors: ((Huntington) OR (Huntingtin Protein)) AND (gene editing). The online tool GeneMania, open access, was also used to analyze probabilities and gene interactions. The silencing of the IT-15 gene causes changes in proteins that interact with the mutated Huntingtin, leading to disturbances in several processes.
Article Details
Authors maintain copyright and grant the HSJ the right to first publication. From 2024, the publications wiil be licensed under Attribution 4.0 International 
 , allowing their sharing, recognizing the authorship and initial publication in this journal.
, allowing their sharing, recognizing the authorship and initial publication in this journal.
Authors are authorized to assume additional contracts separately for the non-exclusive distribution of the version of the work published in this journal (e.g., publishing in an institutional repository or as a book chapter), with acknowledgment of authorship and initial publication in this journal.
Authors are encouraged to publish and distribute their work online (e.g., in institutional repositories or on their personal page) at any point after the editorial process.
Also, the AUTHOR is informed and consents that the HSJ can incorporate his article into existing or future scientific databases and indexers, under the conditions defined by the latter at all times, which will involve, at least, the possibility that the holders of these databases can perform the following actions on the article.
References
van der Burg JM, Björkqvist M, Brundin P. Beyond the brain: widespread pathology in Huntington's disease. Lancet Neurol. 2009;8(8):765-74. https://doi.org/10.1016/S1474-4422(09)70178-4
Martelli A. Aspectos clínicos e fisiopatológicos da doença de Huntington. Arch Health Invest [Internet]. 2014 [cited 2020 22 Sep];3(4):32-39. Avaiable from: http://www.archhealthinvestigation.com.br/ArcHI/article/view/687
Gil-Mohapel JM, Rego AC. Doença de Huntington: uma revisão dos aspectos fisiopatológicos. Rev Neurocienc 2011;19(4):724-34. https://doi.org/10.34024/rnc.2011.v19.8332
Shannon KM. Recent Advances in the Treatment of Huntington's Disease: Targeting DNA and RNA. CNS Drugs. 2020;34(3):219-228. https://doi.org/10.1007/s40263-019-00695-3 PMid:31933283
Intrieri ACU, Filho HB, Sabino MRLS, Ismail M, Furtado CC. Huntington: distúrbio no cromossomo 4. Rev UNILUS Ens Pesq [Internet]. 2015 [cited 2020 Sep 22];12(29):22-34. Avaiable from: http://revista.lusiada.br/index.php/ruep/article/view/687
Kaltenbach LS, Romero E, Becklin RR, et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3(5):e82. https://doi.org/10.1371/journal.pgen.0030082 PMid:17500595 PMCid:PMC1866352
Kolli N, Lu M, Maiti P, Rossignol J, Dunbar GL. CRISPR-Cas9 Mediated Gene-Silencing of the Mutant Huntingtin Gene in an In Vitro Model of Huntington's Disease. Int J Mol Sci. 2017;18(4):754. https://doi.org/10.3390/ijms18040754 PMid:28368337 PMCid:PMC5412339
Gonçalves GAR, Paiva RMA. Terapia gênica: avanços, desafios e perspectivas. Einstein (São Paulo) [Internet]. 2017;15(3):369-75. https://doi.org/10.1590/s1679-45082017rb4024 PMid:29091160 PMCid:PMC5823056
Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010; 38(Suppl 2):W214-20. https://doi.org/10.1093/nar/gkq537 PMid:20576703 PMCid:PMC2896186
Saudou F, Humbert S. The biology of Huntingtin. Neuron. 2016;89(5):910-26. https://doi.org/10.1016/j.neuron.2016.02.003 PMid:26938440
ABH: Associação Brasil Huntington [Internet site]. Contexto genético da DH [access 2020 Nov 12]. Avaiable from: http://abh.org.br/o-que-e-doenca-de-huntington/contexto-genetico-da-dh/
A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72(6):971-83. https://doi.org/10.1016/0092-8674(93)90585-E
Gusella JA, MacDonald ME. Huntington's Disease: seeing the pathogenic process through a genetic lens. Trends Biochem Sci. 2006;31(9):533-40. https://doi.org/10.1186/gm80 PMid:19725930 PMCid:PMC2768966
Landles C, Bates GP. Huntingtin and the molecular pathogenesis of Huntington's Disease. EMBO Rep. 2004;5:958-63. https://doi.org/10.1038/sj.embor.7400250 PMid:15459747 PMCid:PMC1299150
Spitz M. Doença de Huntington e outras coréias. Rev Hosp Universitário Pedro Ernesto. 2010;9(1):29-38.
Frank S. Tetrabenazine: the first approved drug for the treatment of chorea in US patients with Huntington disease. Neuropsychiatric disease and treatment. 2010;6(1):657-65. https://doi.org/10.2147/NDT.S6430 PMid:20957126 PMCid:PMC2951749
Nance MA. Therapy in Huntington's Disease: where are we? Curr Neurol Neurosci Rep. 2012;12(4):359-66. https://doi.org/10.1007/s11910-012-0277-4 PMid:22544535
Shin JW, Kim KH, Chao MJ, et al. Permanent inactivation of Huntington's disease mutation by personalized allele-specific CRISPR/Cas9. Human Molecular Genetics. 2016;25(20):4566-76. https://doi.org/10.1093/hmg/ddw286 PMid:28172889 PMCid:PMC6078600
Ravache M, Abou-Sleymane G, Trottier Y. Neurodegenerative polyglutamine expansion diseases: physiopathology and therapeutic strategies. Pathol Biol (Paris). 2010;58(5):357-66. https://doi.org/10.1016/j.patbio.2009.12.004 PMid:20299163
Sun Y, Savanenin A, Reddy PH, Liu YF. Polyglutamine-expanded huntingtin promotes sensitization of n-methyl-d-aspartate receptors via post-synaptic density 95. J Biol Chem. 2001;276(27):24713-8. https://doi.org/10.1074/jbc.M103501200 PMid:11319238
Li XJ, Li SH, Sharp AH, et al. Huntingtin-associated protein enriched in brain with implications for pathology. Nature. 1995;378(6555):398-402. https://doi.org/10.1038/378398a0 PMid:7477378
Arend MC, Pereira JO, Markorski MM. O Sistema CRISPR/Cas9 e a possibilidade de edição genômica para a cardiologia. Arq Bras Cardiol. 2017;108(1):81-3. https://dx.doi.org/10.5935/abc.20160200
Yang W, Tu Z, Sun Q, Li XJ. CRISPR/Cas9: Implications for modeling and therapy of neurodegenerative diseases. Front Mol Neurosci. 2016;9(30):1-4. https://doi.org/10.3389/fnmol.2016.00030
Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. https://doi.org/10.7554/eLife.00471 PMid:23386978 PMCid:PMC3557905
Savic N, Schwank G. Advances in therapeutic CRISPR/Cas9 genome editing. Transl Res. 2016;168:15-21. https://doi.org/10.1016/j.trsl.2015.09.008 PMid:26470680
Singaraja RR, Huang K, Sanders SS, et al. Altered palmitoylation and neuropathological deficits in mice lacking HIP14. Hum Mol Genet. 2011;20(20):3899-909. https://doi.org/10.1093/hmg/ddr308 PMid:21775500 PMCid:PMC3177655